Introduction to argoFloats
Dan Kelley (https://orcid.org/0000-0001-7808-5911), Jaimie Harbin (https://orcid.org/0000-0003-3774-3732) and Clark Richards (https://orcid.org/0000-0002-7833-206X)
2025-10-30
Source:vignettes/argoFloats.Rmd
argoFloats.RmdAbstract. The argoFloats package makes
it easy identify, download,cache, and analyze oceanographic data
collected by Argo profiling floats, while handling quality control. Our
goal in making this package was to eliminate the gap between the freely
available Argo data and the end user.
Introduction
The argoFloats package provides tools for downloading
and processing Argo profile data. It allows users to focus on core,
biogeochemical (“BGC”) , or deep Argo profiles, and also allows the user
to sift these profiles based on ID, time, geography, variable,
institution, ocean, cycle, direction, profile, dataStateIndicator, etc.
Once downloaded, such data sets can be analyzed within
argoFloats or using other R tools and packages.
Youtube Videos
As an adjunct to the written documentation, the following videos are provided, to introduce concepts and show how to accomplish some every-day tasks. In some cases, sample code is also made available at https://github.com/ArgoCanada/argoFloats/tree/develop/videos.
| Video Name | Creators | Date | URL |
|---|---|---|---|
| argoFloats R 01: Introduction | Dan Kelley & Jaimie Harbin | Apr 9, 2020 | https://youtu.be/xeBoFbb66Nk |
| argoFloats R 02: TS plot near Bermuda | Jaimie Harbin & Dan Kelley | Apr 24, 2020 | https://youtu.be/ZoTrVEMG5Qo |
| argoFloats R 03: New website | Jaimie Harbin & Dan Kelley | Apr 30, 2020 | https://youtu.be/lOvCrRDTmTs |
| argoFloats R 04: Subset by ocean or polygon | Jaimie Harbin & Dan Kelley | May 7, 2020 | https://youtu.be/tcGRB479Udk |
| argoFloats R 05: TS diagram, colour-coded by oxygen | Jaimie Harbin & Dan Kelley | May 14, 2020 | https://youtu.be/Y_SxjcOnW04 |
| argoFloats R 06: Trajectory plot, colour coded by time | Jaimie Harbin & Dan Kelley | May 28, 2020 | https://youtu.be/7BB3UuwjUqo |
| argoFloats R 07: Maps with bathymetry | Jaimie Harbin & Dan Kelley | Jun 4, 2020 | https://youtu.be/Lc32MTMCbbI |
| argoFloats R 08: Introduction to quality control flags | Jaimie Harbin & Dan Kelley | Aug 12, 2020 | https://youtu.be/nN4xs0wCnB4 |
| argoFloats R 09: Advanced Quality Control | Dan Kelley & Jaimie Harbin | Aug 27, 2020 | https://youtu.be/dYzEO5S2GBw |
| argoFloats R 10: Using adjusted data streams | Dan Kelley & Jaimie Harbin | Sep 3, 2020 | https://youtu.be/AuauWeUnopc |
| argoFloats R 11: mapApp() | Dan Kelley & Jaimie Harbin | Sep 21, 2020 | https://youtu.be/PEkIBwXLLpE |
| argoFloats R 12: dataStateIndicator() | Dan Kelley & Jaimie Harbin | Nov 13, 2020 | https://youtu.be/RhLmT5S_XzU |
Preliminary Setup
Since argoFloats is in an active phase of development, it is not yet available on CRAN. Still, it is easily installed in R with
` where, of course, the devtools package must be installed first, if it is not already present on the user’s system.
Typical Work Flow
Figure 1 illustrates the typical workflow with the package, with descriptions of the steps on the left, and names of the relevant functions on the right.
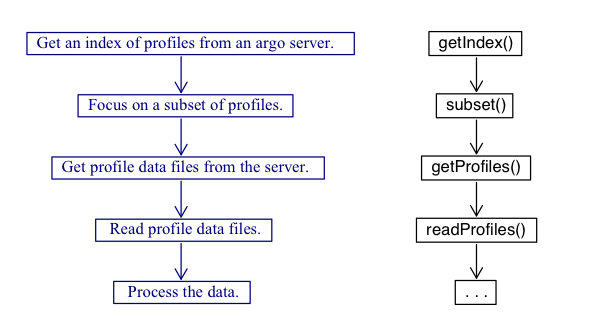
As shown above, the central functions for the argoFloats
package are getIndex(), subset(),
getProfiles(), and readProfiles().
Some built-in data sets are provided for concreteness of illustration
and for testing, but actual work always starts with a call to
getIndex() to download a full index of float data, which we
will demonstrate later in this vignette.
To begin to get familiar with how the argoFloats package
works, we will begin looking at the built in data sets. Built into the
argoFloats package is the index,
indexBgc, indexSynthetic, and
indexDeep indices, referring to core Argo, BGC-Argo, a
combination, and deep Argo respectively. It should be noted that as of
September 21, 2020, indexMerged no longer exists as the
switch to indexSynthetic has been made. For the sake of
this vignette we will focus on the index data set.
The first step is to access the required packages that will be needed during this tutorial, with
To access the embedded index within argoFloats, the
following code is used:
data("index")It is now possible to process the downloaded index using the
argoFloats specialized versions of R “generic” functions,
plot(), [[, summary(), and
show() as shown below.
Processing Steps
The following subsections use built-in data.
Plotting
The specialized plot() command within the
argoFloats package provides simple ways to plot aspects of
argoFloats-class objects. To produce the built in plot and
visualize the coordinates of a section of Argo floats off of the
Bahamas, the following code is used:
plot(index, bathymetry=FALSE) # also, try using bathymetry=TRUE
Figure 2: Built in index demonstrating Argo profiling floats within 200 km of Bahamas.
Extracting Data
Furthermore, the [[ command provides a way to extract
items from argoFloats objects, without getting lost in the
details of storage. For example, if the user wanted to extract the
file within the index data set, instead of
doing index@data$index$file, instead they can simply do
index[["file"]]. (Note that [[<- is
not specialized, since the user is highly discouraged from
altering values within argoFloats objects).
Summarizing
Additionally, the summary() command displays key
features of argoFloats-class objects such as the type,
server, file, URL etc. See summary,argoFloats-method()
further details.
Printing
Lastly, the show() command provides a one-line sketch of
argoFloats-class objects. This gets used by the
print() function. For example if the user types in:
indexThe following output occurs:
argoFloats object of type "index" with 953 itemsHint: This command can be particularly useful when
doing the merge() command, which will be explained in
greater detail further down.
It should be noted that the profile elements within
argoFloats objects are stored as in the form of
argo objects as defined by the oce package.
This means that argoFloats users can rely on a wide variety
of oce functions to analyze their data. The full suite of R
tools is also available, and the vastness of that suite explains why
argoFloats is written in R.
Function Details
getIndex()
Until this point, we have demonstrated how the user can become
familiar with embedded indices. As previously described, however, actual
work always starts with downloading a full index of float data. As shown
by Figure 1, the getIndex() command is used to get an index
of available Argo float profiles, either by downloading information from
a data repository or by reusing an index (stored as an .rda file) that
was prepared by a recent call to the function.
The getIndex() command works by specifying the server,
with first trying the Ifremer server and then the USGODAE server if that
does not work. The next step is to specify the file name. The table
below can be obtained using ?getIndex(). As shown, the user
has the ability to write the specific file name from the server, or to
simply use the embedded nicknames within the package:
"core", "bgc" or "bgcargo", or
"synthetic". The following table summarizes the contents of
the various files indicated by the filename argument.
| File Name | Nickname | Contents |
|---|---|---|
ar_greylist.txt |
- | Suspicious/malfunctioning floats |
ar_index_global_meta.txt.gz |
- | Metadata files |
ar_index_global_prof.txt.gz |
"core" |
Argo data |
ar_index_global_tech.txt.gz |
- | Technical files |
ar_index_global_traj.txt.gz |
- | Trajectory files |
argo_bio-profile_index.txt.gz |
"bgc" or "bgcargo"
|
Biogeochemical data (without S or T) |
argo_bio-traj_index.txt.gz |
- | Biogeochemical trajectory files |
argo_synthetic-profile_index.txt.gz |
"synthetic" |
Synthetic data, successor to "merge"
|
Additionally, the destdir argument has a default of
~/data/argo, where it should be noted that ~
is a short cut for C:\Users\. See ?getIndex()
for further description about the additional arguments for this
command.
To get the index from the Ifremer server, the following code is used:
ai <- getIndex("core")subset()
As shown by Figure 1, the next step when working with the
argoFloats package is to use the subset()
function to focus on a subset of profiles. The argoFloats
package provides tools to sift through profiles based on ID, time,
geography, variable, institution, ocean, dataMode, cycle, direction,
profile, dataStateIndicator, section, etc.
For geographic subsetting, the user has the ability to subset by
circle, rectangle, polygon, or
section.
To subset for specific groups of Argo profiling floats off the coast of Bahamas, the following code is used:
# Subsetting by circle
aiCircle <- subset(ai, circle=list(longitude=-77.5, latitude=27.5, radius=50))
# Subsetting by polygon
lonPoly <- c(-76.5, -76.0, -75.5)
latPoly <- c(25.5, 26.5, 25.5)
aiPoly <- subset(ai, polygon=list(longitude=lonPoly, latitude=latPoly))
# Plotting the subsets together
CP <- merge(aiCircle, aiPoly)
plot(CP, bathymetry=FALSE) # also, try using bathymetry=TRUE
Figure 3: 50 km radius and polygon subset of Argo profiling floats found off the coast of Bahamas
Exercise 1: Use the subset by rectangle function to add a rectangle subset onto Figure 3 and subset all of the data to only include samples between 2012 and 2020.
As of March, 2021, a subset by section was developed to
create a section of Argo data, similarly to what is done with CTD data.
A subset by section combined with subset by
time can provide insightful information of Argo vs CTD
sampling efforts. An example of this is highlighted below, which
compares a section of Argo data from the Mediterranean outflow region
across to North America to the line A03 CTD section data collected in
1993-09-11.
Exercise 2: Use the information from Figure 4 to compare Argo and CTD section sampling methods.
library(oce)
library(argoFloats)
oldpar <- par(no.readonly=TRUE)
par(mfrow=c(2,1))
data(section, package="oce")
#getIndex()
ai <- getIndex()
#subset by section
lonlim <- c(-70, -64,-10)
latlim <-c(40,35,35)
index1 <- subset(ai, section=list(longitude=lonlim, latitude=latlim, width=100))
#subset by time
from <- as.POSIXct("2020-09-23", tz="UTC")
to <- as.POSIXct("2020-10-25", tz="UTC")
index2 <- subset(index1, time=list(from=from, to=to))
plot(index2, bathymetry=FALSE, asp=1/cos(mean(range(unlist(index2[["latitude"]]), na.rm=TRUE))*pi/180), mgp=getOption("oceMgp")
)
points(lonlim, latlim, pch=21, col="black", bg="red", type="o")
plot(section, which="map", col="tan")
par(oldpar)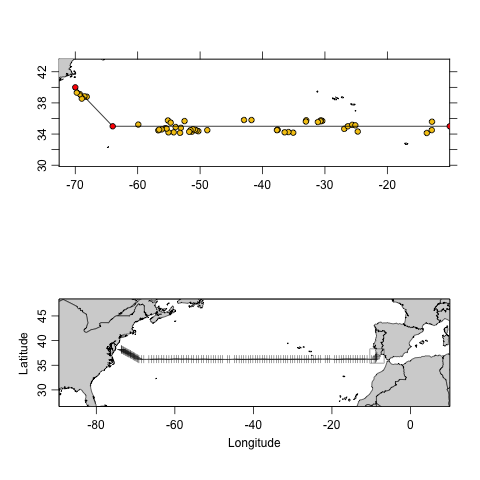
getProfiles()
The next step within the argoFloats package is the
getProfiles() function. This takes an index constructed
with getIndex(), possibly after focusing with
subset,argoFloats-method(), and creates a list of files to
download from the server named in the index. Then these files are
downloaded to the destdir directory, using filenames
inferred from the source filenames. The value returned by
getProfiles() is suitable for use by
readProfiles() function.
readProfiles()
The readProfiles() command works with either a list of
local NetCDF files, or a argoFloats object type
profiles, as created by getProfiles().
The command can be useful for analyzing individual profiles, for example:
index1 <- subset(index, 1:2) # To subset for profiles
profiles <- getProfiles(index1)
argos <- readProfiles(profiles)
argosClean <- applyQC(argos)
plot(argosClean, which="profile", type="p")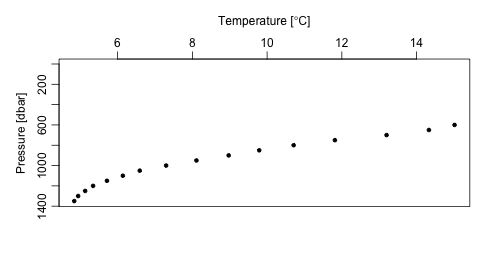
Exercise 3: Using the profile in the previous example, plot a TS diagram.
Appendix 1: Work Flow With Local Index Files
In normal usage, getIndex() works by downloading a
gzipped CSV file (with suffix .gz) from an Argo server,
analyzing that file writing an R Data Archive file (with suffix
.rda), and then deleting the .gz file.
However, if getIndex() is called with the parameter
keep set to TRUE, it will retain the .gz file
in the directory named by the destdir argument. This can be
useful if there is a need to work with the data in a processing system
other than argoFloats.
It is possible to instruct getIndex() to skip remote
downloads, to instead use a local .gz or .rda
file. This is done by providing the file name as the first argument, and
by setting server=NULL, e.g. using
index <- getIndex("~/data/argo/ar_index_global_prof.txt.gz", server=NULL)or
index <- getIndex("~/data/argo/ar_index_global_prof.rda", server=NULL)after which the other processing steps, as outlined in the previous section, can be followed.
The .rda approach is better than the .gz
one, for two reasons: (1) it is faster and (2) it usually sets a better
foundation for later calls to getProfiles(). The second
factor is a result of the fact that the .gz files on
servers do not contain an indication of the server URL, whereas the
locally-generated .rda files do so. (Recall that
getIndex() can try a list of servers, and it records in the
.rda file the first one that yielded results from a
download attempt.)
Whether the .gz or .rda method is chosen,
it is worth noting that index files go out of date, as new data become
available, and also as pointers to real-time data are made obsolete
because of replacement by delayed-mode equivalents. Even so, it can be a
good idea to download an index file before a classroom/workshop exercise
and to share it over a local network, avoiding placing too much stress
on the larger network.
Solutions to Exercises
Exercise 1: Use the subset by rectangle function to add a rectangle subset onto Figure 3 and subset all of the data to only include data between 2012 and 2020.
library(argoFloats)
ai <- getIndex()
# Subset by circle
index1 <- subset(ai, circle=list(longitude=-77.5, latitude=27.5, radius=50))
# Subset by polygon
lonPoly <- c(-76.5, -76.0, -75.5)
latPoly <- c(25.5, 26.5, 25.5)
index2 <- subset(ai, polygon=list(longitude=lonPoly, latitude=latPoly))
# Subset by rectangle
lonRect <- c(-76.5, -76)
latRect <- c(27, 28)
index3 <- subset(ai, rectangle=list(longitude=lonRect, latitude=latRect))
# Merge the subsets together
index4 <- merge(index1, index2)
index5 <- merge(index3, index4) # Note right now can only merge 2 indices together
# Subset for year 2012-2020
index6 <- subset(index5, time=list(from="2012-01-01", to="2020-01-01"))
# Plot data
plot(index6, bathymetry=FALSE) # also, try using bathymetry=TRUEExercise 2: Use the information from Figure 4 to compare Argo and CTD section sampling efforts.
Figure 4 reveals that it takes almost 3 months of Argo sampling to reproduce a 1 month CTD section cruise. Additionally, it displayed that there are geographical gaps within the Argo data. On the other hand, it should be noted that by extending the latitude range, these gaps are quickly filled in. In conclusion, Argo sampling is much more cost effective, and it can allow for collection of more information (i.e. nutrients, oxygen, backscattering, etc.) if such thing is desired.
Exercise 3: Using the profile in the previous example, plot a TS diagram.
library(argoFloats)
ai <- getIndex("synthetic")
sub <- subset(ai, 1:2) # To subset for profiles
profiles <- getProfiles(sub)
argos <- readProfiles(profiles)
argosClean <- applyQC(argos)
plot(argosClean, which="TS")